File Input and Output
- How are files stored?
- Reading a text file, line-by-line
- File Object
- Lists from files
- Dictionaries from files
- Exercise: Find the tallest tree
- Writing a text file
Other Tools for Python File I/O
first, make sure you are in the right directory
you should see animals.txt and README.txt
In [1]:
pwd
Out[1]:
In [2]:
ls
In [3]:
cat README.txt
In [4]:
cat animals.txt
1. How are files stored?
Binary Encoding of Text
A simple plain text file that contains the plain-text string "Rockefeller U." is stored in 15 bytes as:
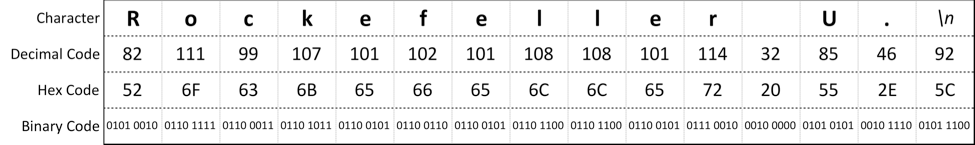
RU
Occasional Pain Point: \n versus \r\n
\n= New Line- In most Unix systems including Mac OSX,
\nis the standard line terminator for text files - The non-printable value, 0101 1100, will occupy the byte that signifies the new line marker
- In most Unix systems including Mac OSX,
\r= Carriage Return- Windows text files typically terminate lines with the sequence
\r\n - Whle Python does its best to insulate you from having to keep track of
\nversus\r\n, you may come across situations where the presence of Carriage Return characters is an issue.
- Windows text files typically terminate lines with the sequence
Where does "Carriage Return" come from anyway?

CRLF
Common Plain-Text file types
- Text (.txt)
- Comma-Separated Values (.csv)
- Most spreadsheets allow for export of .csv files
- Common way of sharing "Flat" flies (one row = one observation)
- Many possible variations: headers, row names, separator characters, quotes, etc.
- Space-padded, tab-delimited (), etc.
- Source code, HTML, "Natural Language," XML, JSON, Yaml, etc. etc.
Files with Other Binary Encodings
- Documents: databases (many open and proprietary formats), .pdf, .doc, .xls, etc.
- Images: .jpg, .png, many other formats
- Audio: mp3, ogg, etc.
- Data from sensors, repeated measures, archaic formats, etc.
The Point:
- Find out what format your input data is in
- To the extent possible, look at your data
- Linux utilities:
cat,lesshead,tail,wc, etc.
- Linux utilities:
- Test your expectations
- Decide how to handle missing data and "bad" data
- Big subjects, not covered today
- Allow plenty of time for dealing with file format ideosyncracies and conversions
2. Reading a text file, line-by-line
- Look at the data
- Read into a File Object
- Create a List from the File
- Create a Dictionary from File
Animals dataset
A list of animals, their body mass, and their brain mass
Question of interest: Which animal has the largest brain-to-body mass ratio? And the smallest?
Examine the data
Here we use the iPython Notebook "magics" to call the shell. You can also just do this from the command line
- Look at the data
- Read the docs
In [5]:
%%bash
ls
In [6]:
%%bash
wc Animals.txt
# what does the output mean? call: man wc
In [7]:
%%bash
head Animals.txt
Read the data into a file object
- two approaches
In [8]:
# For convenience, assign the path and file name to variables
read_this = 'Animals.txt'
print read_this
In [9]:
# Approach 1: open, read, close
file_in = open(read_this, 'r')
all_lines = file_in.readlines()
file_in.close() # important to close the file
In [9]:
In [10]:
print "Name of the file: ", file_in.name
print "Is File Closed? : ", file_in.closed
print "Opening mode : ", file_in.mode
print "Softspace flag : ", file_in.softspace
Another way to read a file: python with()
- This formulation will automatically close the file after reading it
In [11]:
with open(read_this, 'r') as file_in:
lines = file_in.readlines()
# file_in.close()
In [12]:
type(lines)
Out[12]:
In [13]:
lines[0:5]
Out[13]:
In [14]:
# print each individual line -- iterate over the file
for waffles in lines:
print waffles
In [15]:
# repr := representation of the line, revealing hidden characters
for line in lines:
print repr(line)
Splitting Up Each Line: From a String to a List
In [16]:
some_line = lines[2]
print some_line
In [17]:
some_line[0]
Out[17]:
In [18]:
type(some_line)
Out[18]:
In [18]:
In [19]:
# What is the first element of some_line?
some_line[0]
# is that what you expected?
Out[19]:
In [20]:
for _ in some_line:
print _
In [21]:
print type(some_line)
In [22]:
line_split_at_commas = some_line.split(",") # forms a python List object, splitting at the comma characters
print line_split_at_commas
In [23]:
print some_line.strip() # gets rid of whitespace...
# print type(some_line.strip()) # though result is still a string
In [24]:
my_new_line = some_line.strip()
print my_new_line[0]
In [24]:
In [24]:
In [25]:
line_list = some_line.strip().split(",")
print line_list
In [26]:
for l in line_list:
print l
In [27]:
3 + 3
Out[27]:
In [30]:
'3' + '3'
Out[30]:
Multiple assignment trick: list unpacking
In [31]:
animal, body_mass, brain_mass = line_list
print "Animal = ", animal
print "Body Mass =", float(body_mass)
print "Brain Mass =", float(brain_mass)
3. Exercise: Which animal has biggest brain-to-body ratio? Smallest?
- Read Animal.txt into a file object
- Calculate brain / body ratio
- pay attention to integer arithmetic issues
- Print ratio for each animal
- Find min and max.
Do your results make sense to you?
- Bonus 1:
- Create variables to keep track of min and max
- Just print the animal with the max and the animal with the min
- Bonus 2:
- Find average ratio
In [31]:
pseudo-code
- read the file into a LIST!!!!!!! here it happens to be called "lines"
- for item in lines:
- clean up the item. i,e., convert the item from a string into a NEW list
- figure out which elements are the numbers you want
- take the ratio
- print the animal name and the ratio
In [31]:
In [32]:
# List-based solution
with open(read_this, 'r') as file_in:
lines = file_in.readlines()
ratios = []
for line in lines:
clean_line = line.strip().split(',')
animal, body_mass, brain_mass = clean_line
if animal == "Animal":
pass # skip the header line
else:
ratio = float(brain_mass) / (1000 * float(body_mass))
print "Animal=",animal, "\tratio=", ratio
ratios.append([animal, ratio])
In [33]:
# dictionary-based solution
with open(read_this, 'r') as file_in:
lines = file_in.readlines()
ratios_dict = {} # empty dictionary
for line in lines:
clean_line = line.strip().split(',')
animal, body_mass, brain_mass = clean_line
if animal == "Animal":
pass # skip the header line
else:
ratios_dict[animal] = float(brain_mass) / (1000 * (float(body_mass))) # WATCH parentheses and order of operations
In [34]:
print ratios_dict
In [35]:
print ratios_dict.keys()
In [36]:
print ratios_dict.values()
In [37]:
print (ratios_dict['Cat'])
In [38]:
# using the dictionary like a database
my_study_subjects = ['Cat', 'Rabbit', 'Goat']
for subj in my_study_subjects:
print subj, (ratios_dict[subj])
In [38]:
4. Writing a text file
- Suppose we want to save our calculated brain-to-body ratios
In [172]:
file_out_name = 'Animal_brain_to_body_ratio.txt'
file_out = open(file_out_name, 'w')
for r in ratios:
line_out = r[0] + ", " + str(r[1]) + "\n"
file_out.write(line_out)
file_out.close()
In [172]:
5. Other Tools for Reading and Writing Files
csv module
- somewhat more "helpful" way of reading and writing flat files
- official documentation
pandas
- Beyond file i/o, a powerful set of full-fledged "Data Wrangling" tools for python
- cross-sectional, time-series
- Intro video at http://vimeo.com/59324550
- Book info: http://shop.oreilly.com/product/0636920023784.do
Serialization
- Text files such as .txt and .csv are useful for recording "flat" files
- However sometimes we want to save more complex objects, like dictionaries
- This is referred to as
serialization - Many serialization tools in python, including "pickle"
In []:
In []: